Microbiome
 Fecal froths in the David Lab at Duke University
Fecal froths in the David Lab at Duke UniversityA microbiome is the metagenome of all the microorganisms in a particular ecology: your gut, your right hand, your left hand, an insect’s digestive tract, the flour you buy at a grocery store, and just about every surface on Earth. Microbiome science has exploded recently, especially with the NIH Human Microbiome Project established in 2008. With this research, a more holistic, interdependent picture of life has been developing.
In the spring term of 2019, the S-1 Lab visited Duke University’s David Lab . Lawrence David, PhD, and his team have been investigating how the human gut microbiome differs across individuals over time with changes in diet as well as how those differences impact other elements in human health and cognition. PhD student Zachary Holmes gave us a tour, which sparked a series of engagements with microbiome science. This culminated in a pair of panels organized for the 2019 Annual Meeting of the Society for Literature, Science, and the Arts (SLSA), and which were in turn part of a larger stream of panels devoted to the microbiome.
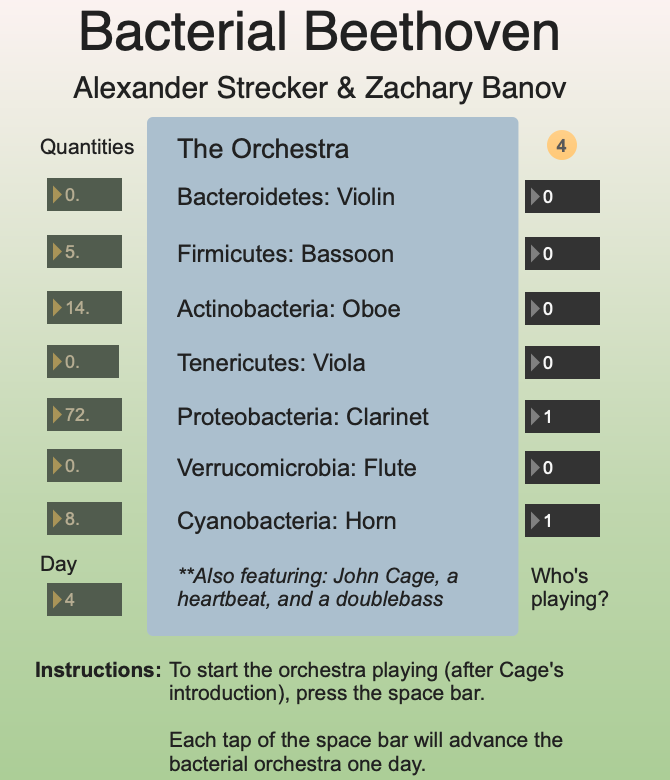
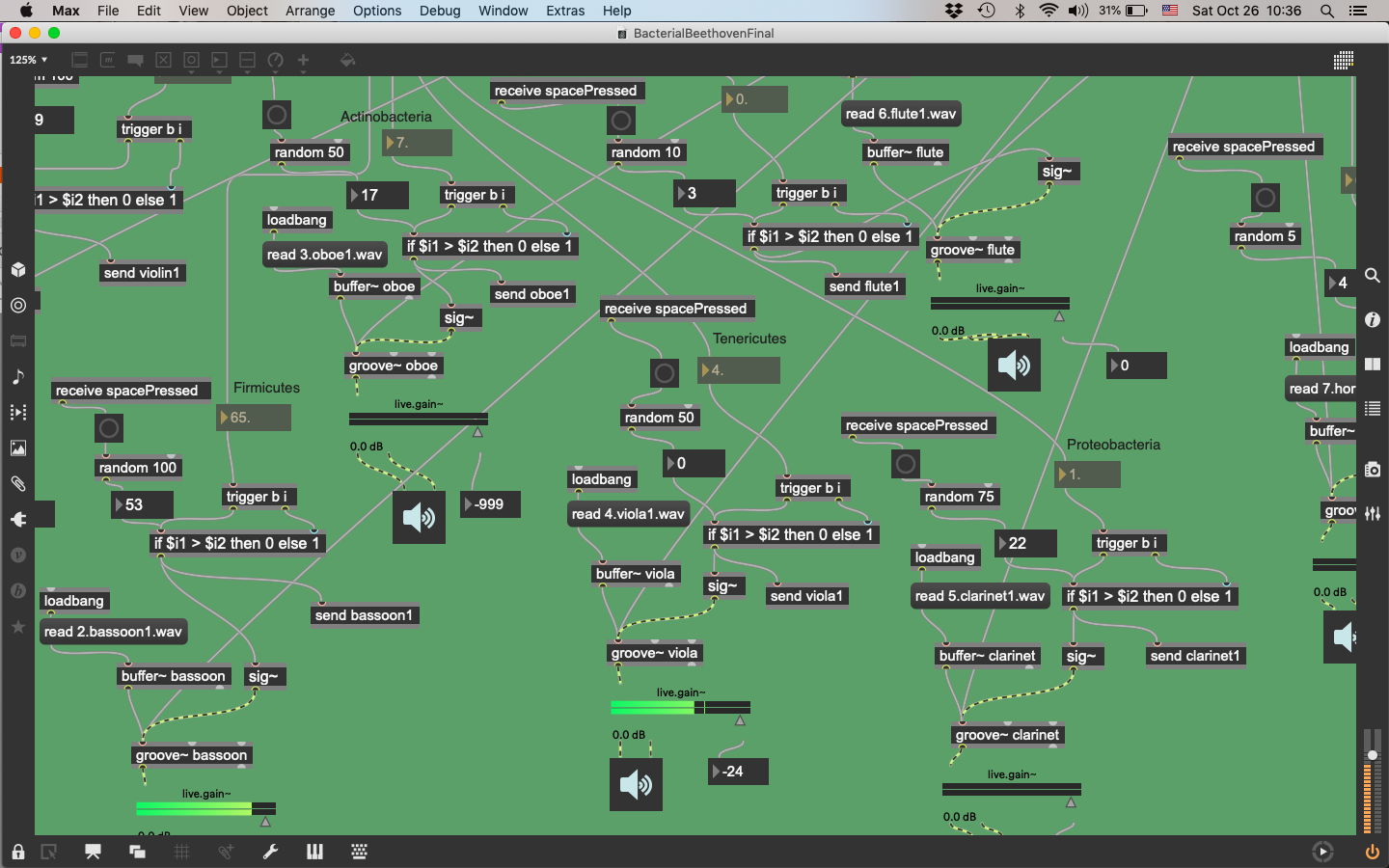
One project, created by S-1 member Alex Strecker and former MA student Zachar Banov, transformed the genomic data sequenced from one year of daily stool samples into an electronic music piece. Each bacterial genus “plays” an element in the orchestra based on their relative population. Strecker and Banov’s creation explores how such an unwieldy dataset must be processed so that it may present sensible information to us. Above is a screenshot of the interface when playing. Below is a screenshot of the arrangement made using Max .
Other presentations discussed research into machine learning, the commercialization of the microbiome within the health industry, Hegel’s penchant for alimentary processes, and what Simondon’s philosophy of individuation might tell us about the changes to the gut microbiome that takes place when refugees are forced to relocate.
For a full list of abstracts, see the “Microbial Futures” panels in the SLSA Program .